BIOIMAGERY
Keen Eye AI
Plateform
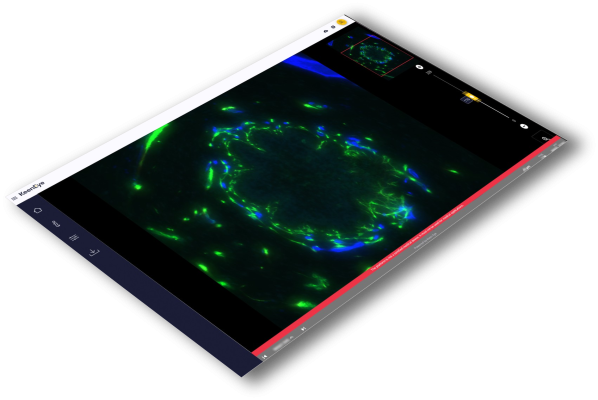
The Keen Eye Platform is a web-based plateform which can be used to visualize different types of bioimagery (TMA, whole slide, H&E, IHC, multiplex, fluorescent, bright-field, 2D, 3D and more). It is used by the HUGODECA consortium to store and visualize in a fluid manner 2D and 3D image stacks, all as well as ISS cell maps with up to 10 million annotations on one image.
Authentication required
GENE EXPRESSION
genoViewer
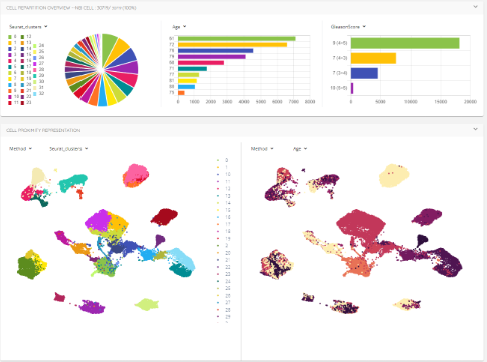
The genoViewer can be used to explore and analyze single-cell or spatial transcriptomics datasets. It enables you to view the repartition of cells according to multiple criteria and to compare gene expression in human developmental cells.
OMICS DATA & BIOIMAGERY
Scripts
& Pipelines
developped by the HuGoDeCA consortium
Protocols
applied to prepare the samples
Clearing techniques have been developed to transparentize mouse brains, thereby preserving 3D structure, but their complexity has limited their use. Here, we show that immunolabeling of axonal tracts followed by optical clearing with solvents (3DISCO) and light-sheet microscopy reveals brain connectivity in mouse embryos and postnatal brains. We show that the Robo3 receptor is selectively expressed by medial habenula axons forming the fasciculus retroflexus (FR) and analyzed the development of this commissural tract in mutants of the Slit/Robo and DCC/Netrin pathways. Netrin-1 and DCC are required to attract FR axons to the midline, but the two mutants exhibit specific and heterogeneous axon guidance defects. Moreover, floor-plate-specific deletion of Slit ligands with a conditional Slit2 allele perturbs not only midline crossing by FR axons but also their anteroposterior distribution. In conclusion, this method represents a unique and powerful imaging tool to study axonal connectivity in mutant mice.
Morgane Belle, David Godefroy, Chloé Dominici, Céline Heitz-Marchaland, Pavol Zelina, Farida Hellal, Frank Bradke, Alain Chédotal
DOI : https://doi.org/10.1016/j.celrep.2014.10.037 LICENCE : CC BY 3.0
Morgane Belle, David Godefroy, Chloé Dominici, Céline Heitz-Marchaland, Pavol Zelina, Farida Hellal, Frank Bradke, Alain Chédotal
DOI : https://doi.org/10.1016/j.celrep.2014.10.037 LICENCE : CC BY 3.0
Protocol for enrichment of fetal gonadal cells
Regina Hoo, Roser Vento-Tormo, Carmen Sancho
DOI : dx.doi.org/10.17504/protocols.io.66fhhbn LICENCE : CC BY 4.0
Regina Hoo, Roser Vento-Tormo, Carmen Sancho
DOI : dx.doi.org/10.17504/protocols.io.66fhhbn LICENCE : CC BY 4.0
Current long read sequencing, e. g. PacBio and Nanopore, requires high molecular weight (HMW) and highly pure DNA. Many fungal and plant species have high content of polysaccharides and other contaminants that are co-precipitated with DNA during ethanol precipitation. This protocol aims to purify HMW DNA from the DNA solution obtained using Benjamin Schwessinger’s protocol. The protocol was tested fuccessfully in wheat leaf rust, barley leaf rust, wheat stem rust and myrtle rust. Critcal step to seperate HMW DNA from low molecular weight DNA and other contaminants during ethanol precipitation is to allow cotton-fiber-like HMW DNA to sediment by gravity, while other small molecule DNA and contaminants remain in the supernatant.
DOI : dx.doi.org/10.17504/protocols.io.66fhhbn LICENCE : CC BY 4.0
